class: center, middle # Point Process Analysis ### Ignacio Sarmiento-Barbieri --- # Tipos de Datos Espaciales Los datos espaciales vienen en muchas "formas" y "tamaños", los tipos más comunes de datos espaciales son: - **Punto** - **Líneas** - **Polígonos** - **Grillas (*o raster*)** --- # The earth ain't flat <img src="figures/world-1.png" width="800" style="display: block; margin: auto;" /> --- # Projections - Geographic coordinate systems: coordinate systems that span the entire globe (e.g. latitude / longitude). - Projected coordinate systems: coordinate systems that are localized to minimize visual distortion in a particular region (e.g. Robinson, UTM, State Plane) --- # Projections <img src="figures/proj.png" width="800" style="display: block; margin: auto;" /> --- # Which projection should I choose? - “There exist no all-purpose projections, all involve distortion when far from the center of the specified frame” (Bivand, Pebesma, and Gómez-Rubio 2013) - In some cases, it is not something that we are free to decide: “often the choice of projection is made by a public mapping agency” (Bivand, Pebesma, and Gómez-Rubio 2013). - This means that when working with local data sources, it is likely preferable to work with the CRS in which the data was provided. - For Bogotá the IGAC promotes the adoption of MAGNA-SIRGAS. EPSG code: 4626 --- ## Reading Spatial Data in R Packages: ```r require("sf") ``` ``` ## Loading required package: sf ``` ``` ## Linking to GEOS 3.10.2, GDAL 3.4.2, PROJ 8.2.1; sf_use_s2() is TRUE ``` ```r require("tidyverse") require("here") ``` ``` ## Loading required package: here ``` ``` ## here() starts at /Users/ignaciosarmiento/Dropbox/Teaching/2022/SpatialDataEng/Lectures/Lecture2 ``` --- ## Reading Spatial Data in R ```r bars<-st_read(here("egba/EGBa.shp")) ``` ``` ## Reading layer `EGBa' from data source ## `/Users/ignaciosarmiento/Dropbox/Teaching/2022/SpatialDataEng/Lectures/Lecture2/egba/EGBa.shp' ## using driver `ESRI Shapefile' ## Simple feature collection with 515 features and 7 fields ## Geometry type: POINT ## Dimension: XY ## Bounding box: xmin: -74.17607 ymin: 4.577897 xmax: -74.02929 ymax: 4.806253 ## Geodetic CRS: MAGNA-SIRGAS ``` --- ## Visualizing Points .pull-left[ ```r ggplot()+ geom_sf(data=bars) + theme_bw() + theme(axis.title =element_blank(), panel.grid.major = element_blank(), panel.grid.minor = element_blank(), axis.text = element_text(size=6)) ``` ] .pull-right[ 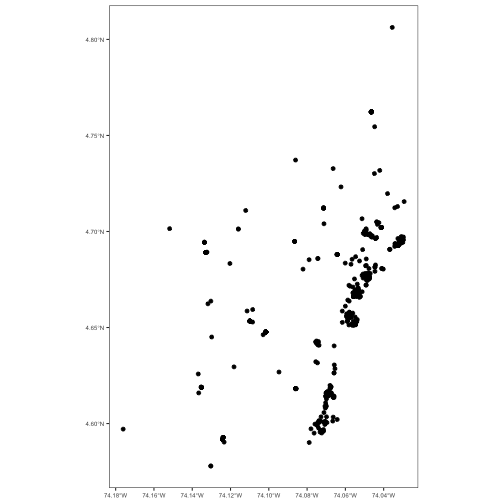<!-- --> ] --- class: center, middle # Point Process Methods --- # Questions - Are the points spread uniformly over the survey region? - Are the points randomly scattered? - Is there evidence of clustering? - Does the density of points depend on an explanatory variable? -- To give a sensible answer we must recognise that these questions are not about the points themselves, but about the way the points were generated. --- <img src="figures/point_realizations.png" width="900" height="600" style="display: block; margin: auto;" /> --- # Point Process and Notation <img src="figures/fig52.png" width="400" height="400" style="display: block; margin: auto;" /> --- # Complete spatial randomness (CSR) Properties: - homogeneity - independence --- # CSR: homogeneity <img src="figures/fig56.png" width="2456" height="400" style="display: block; margin: auto;" /> --- # CSR: independence <img src="figures/fig57.png" width="2200" height="400" style="display: block; margin: auto;" /> --- # Simulation of CSR ```r require("spatstat") ``` ``` ## Loading required package: spatstat ``` ``` ## Loading required package: spatstat.data ``` ``` ## Loading required package: spatstat.geom ``` ``` ## spatstat.geom 2.4-0 ``` ``` ## ## Attaching package: 'spatstat.geom' ``` ``` ## The following object is masked from 'package:scales': ## ## rescale ``` ``` ## Loading required package: spatstat.random ``` ``` ## spatstat.random 2.2-0 ``` ``` ## Loading required package: spatstat.core ``` ``` ## Loading required package: nlme ``` ``` ## ## Attaching package: 'nlme' ``` ``` ## The following object is masked from 'package:dplyr': ## ## collapse ``` ``` ## Loading required package: rpart ``` ``` ## spatstat.core 2.4-4 ``` ``` ## Loading required package: spatstat.linnet ``` ``` ## spatstat.linnet 2.3-2 ``` ``` ## ## spatstat 2.3-4 (nickname: 'Watch this space') ## For an introduction to spatstat, type 'beginner' ``` --- class: center # Simulation of CSR ```r set.seed(10101) plot(rpoispp(50)) ``` <!-- --> --- class: center # Simulation of CSR ```r plot(rpoispp(100,nsim=6)) ``` <!-- --> --- # Inhomogeneous Poisson process .pull-left[ ```r lambda <- function(x,y) { 100 * (x^2+y) } X <- rpoispp(lambda, win=square(1)) ``` ] .pull-right[ ```r plot(X) ``` <!-- --> ] --- class: center, middle # Exploratory Data Analysis --- # Intensity ```r plot(swedishpines, main = "") ``` <!-- --> --- # Intensity ```r npoints(swedishpines)/area(Window(swedishpines)) ``` ``` ## [1] 0.007395833 ``` ```r intensity(swedishpines) ``` ``` ## [1] 0.007395833 ``` ```r unitname(swedishpines) ``` ``` ## 0.1 metres ``` --- # Estimating Homogeneous Intensity ```r summary(swedishpines) ``` ``` ## Planar point pattern: 71 points ## Average intensity 0.007395833 points per square unit (one unit = 0.1 metres) ## ## Coordinates are integers ## i.e. rounded to the nearest unit (one unit = 0.1 metres) ## ## Window: rectangle = [0, 96] x [0, 100] units ## Window area = 9600 square units ## Unit of length: 0.1 metres ``` --- # Estimating Homogeneous Intensity ```r intensity(rescale(swedishpines)) ``` ``` ## [1] 0.7395833 ``` --- # Estimating Homogeneous Intensity ```r X <- rescale(swedishpines) lam <- intensity(X) (sdX <- sqrt(lam/area(Window(X)))) ``` ``` ## [1] 0.08777239 ``` --- # Quadrant Counting ```r swp <- rescale(swedishpines) Q3 <- quadratcount(swp, nx=3, ny=3) Q3 ``` ``` ## x ## y [0,3.2) [3.2,6.4) [6.4,9.6] ## [6.67,10] 8 6 7 ## [3.33,6.67) 8 11 9 ## [0,3.33) 5 6 11 ``` --- # Quadrant Counting <!-- --> --- # Quadrant Counting test for homogeneity ```r intensity(Q3) ``` ``` ## x ## y [0,3.2) [3.2,6.4) [6.4,9.6] ## [6.67,10] 0.75000 0.56250 0.65625 ## [3.33,6.67) 0.75000 1.03125 0.84375 ## [0,3.33) 0.46875 0.56250 1.03125 ``` --- # Quadrant Counting test for homogeneity ```r tS <- quadrat.test(swp, 3,3) tS ``` ``` ## ## Chi-squared test of CSR using quadrat counts ## ## data: swp ## X2 = 4.6761, df = 8, p-value = 0.4169 ## alternative hypothesis: two.sided ## ## Quadrats: 3 by 3 grid of tiles ``` ```r tS$p.value ``` ``` ## [1] 0.4168565 ``` --- # Quadrant Counting test for homogeneity <!-- --> --- # KDE Sean `\((x_1, x_2, \cdots, x_n) \sim iid\)` from some unknown `\(f\)`, we want to estimate it `$$\hat{f_h}(x)=\frac{1}{nh}\sum_{i=1}^nK\left(\frac{x-x_i}{h}\right)$$` --- # KDE <img src="figures/fig1.png" width="500" height="400" style="display: block; margin: auto;" /> --- # KDE Bandwidth `\begin{align} MSE (\hat{f}(x)) = \left( Sesgo (\hat{f}(x))\right)^2 + Var (\hat{f}(x)) \end{align}` La expresión aproximada en `\(x_o\)` está dada por `\begin{align} Sesgo [\hat{f} (x_o)] \approx \frac{h^2}{2} f''(x_o ) \int_{-\infty}^\infty K(\phi) \phi^2 d\phi \end{align}` `\begin{align} Var [\hat{f} (x_o)] \approx \frac{1}{n\times h} f(x_o ) \int_{-\infty}^\infty K^2(\phi) d\phi \end{align}` --- # KDE <img src="figures/fig2.png" width="500" height="400" style="display: block; margin: auto;" /> --- # KDE <img src="figures/fig3.png" width="500" height="400" style="display: block; margin: auto;" /> --- # KDE Bandwidth - Scott's Rule: `\(h \approx 1.06⋅\hat{\sigma} n^{−1/5}\)`. - Silverman: `\(h \approx 0.9 min \left\{\hat{\sigma}, \frac{IQR}{1.35}\right\} n^{−1/5}\)`. - Cross-Validation: `\begin{align} CV_k(h) = \frac{1}{n} \sum_{i=1}^n \log \hat{f}_{-k}(X_k) \end{align}` --- # KDE <img src="figures/fig4.png" width="500" height="400" style="display: block; margin: auto;" /> --- ### KDE: Kernel Properties 1. `\(K(u)=K(-u)\)` 2. `\(\int_{-\infty}^\infty K(u) du = 1\)` 3. `\(K'(u)<0\)` when `\(u>0\)` 4. `\(E[K]=0\)` --- ### KDE: Kernel <img src="figures/fig5.png" width="500" height="400" style="display: block; margin: auto;" /> --- ### KDE: Kernel <img src="figures/fig6.png" width="500" height="400" style="display: block; margin: auto;" /> --- ### KDE: Kernel <img src="figures/fig7.png" width="500" height="400" style="display: block; margin: auto;" /> --- ### KDE: Kernel <img src="figures/fig8.png" width="500" height="400" style="display: block; margin: auto;" /> --- ### KDE: Kernel Kernel functions: - Gaussian - Epanechnikov - Rectangular - Triangular --- ### KDE: Kernel <img src="figures/fig9.png" width="500" height="400" style="display: block; margin: auto;" /> --- ### KDE: Kernel ```r db<-data.frame(place=c("CBD"),lat=c(4.651887), long=c(-74.05812)) db<-st_as_sf(db,coords=c('long','lat'),crs=4326) #EPSG:4326 - WGS 84, latitude/longitude coordinate system db<-st_transform(db, 4686) z<-st_distance(bars,db) head(z) ``` ``` ## Units: [m] ## [,1] ## [1,] 4330.199 ## [2,] 4330.199 ## [3,] 4289.943 ## [4,] 4372.001 ## [5,] 4291.624 ## [6,] 4403.613 ``` --- ### KDE: Kernel ```r z<-units::set_units(z,"km") head(z) ``` ``` ## Units: [km] ## [,1] ## [1,] 4.330199 ## [2,] 4.330199 ## [3,] 4.289943 ## [4,] 4.372001 ## [5,] 4.291624 ## [6,] 4.403613 ``` --- ### KDE: Kernel ```r plot(density(z)) ``` <!-- --> --- ### KDE: Kernel ```r plot(density(z,bw="nrd0", kernel='gaussian')) ``` 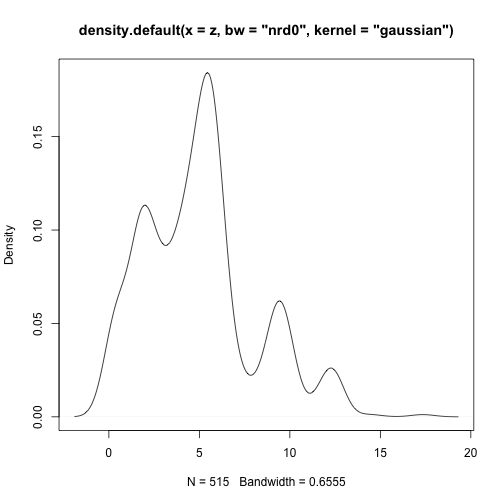<!-- --> --- ### KDE: Kernel ```r plot(density(z,bw="nrd0", kernel='epanechnikov')) ``` 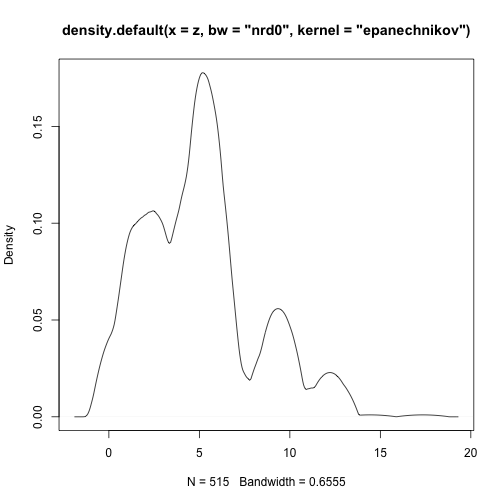<!-- --> --- ### KDE: Kernel ```r plot(density(z,bw="nrd0", kernel='rectangular')) ``` 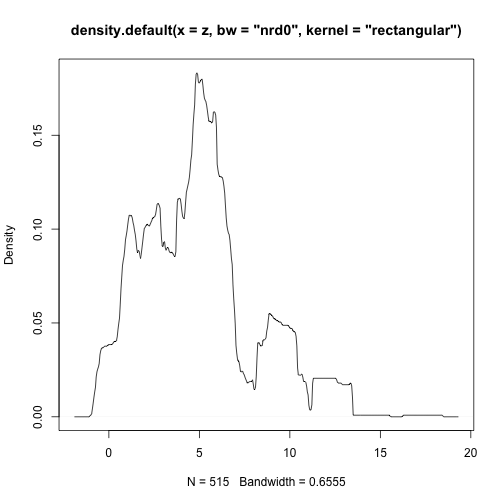<!-- --> --- ### Bivariate KDE `\begin{align} \hat{f}(x;H):=\frac{1}{n|H|^{1/2}}\sum_{i=1}^nK\left(H^{-1/2}(\mathbf{x}-\mathbf{x}_i)\right) \end{align}` `\begin{align} \hat{f}(x,y) = \frac{1}{nh_xh_y}\sum_{i=1}^n K\left(\frac{X_i-x}{h_x}\right)K\left(\frac{Y_i-x}{h_y}\right) \end{align}` --- ### Bivariate KDE .pull-left[ ```r ggplot(bars, aes( x=LONGITUD, y=LATITUD) ) + geom_density_2d(size = 0.25, colour = "black") + theme_bw() ``` ] .pull-right[ 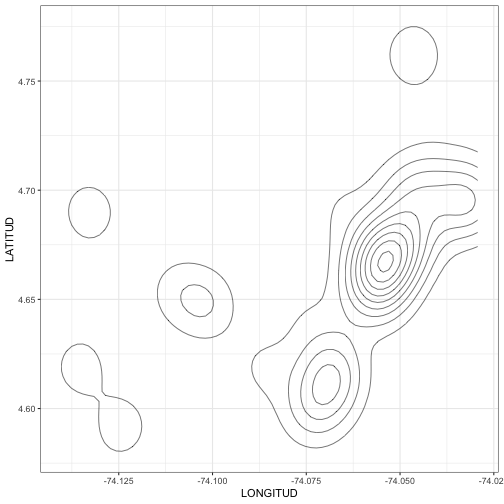<!-- --> ] --- ### Bivariate KDE ```r require("SpatialKDE") bars<-st_transform(bars,21818) cell_size <- 2000 band_width <- 1000 grid_bars <- bars %>% create_grid_rectangular(cell_size = cell_size, side_offset = band_width) ``` --- ### Bivariate KDE ```r kde <- bars %>% kde(band_width = band_width, kernel = "epanechnikov", grid = grid_bars) kde ``` ``` ## Simple feature collection with 100 features and 1 field ## Geometry type: POLYGON ## Dimension: XY ## Bounding box: xmin: 590396.2 ymin: 505070.2 xmax: 610396.2 ymax: 533070.2 ## Projected CRS: Bogota 1975 / UTM zone 18N ## First 10 features: ## geometry kde_value ## 1 POLYGON ((590396.2 505070.2... 0.0000000 ## 2 POLYGON ((592396.2 505070.2... 0.0000000 ## 3 POLYGON ((594396.2 505070.2... 0.0000000 ## 4 POLYGON ((596396.2 505070.2... 0.2990913 ## 5 POLYGON ((598396.2 505070.2... 0.0000000 ## 6 POLYGON ((600396.2 505070.2... 0.0000000 ## 7 POLYGON ((602396.2 505070.2... 0.0000000 ## 8 POLYGON ((590396.2 507070.2... 0.9865835 ## 9 POLYGON ((592396.2 507070.2... 0.0000000 ## 10 POLYGON ((594396.2 507070.2... 0.0000000 ``` --- ### Bivariate KDE .pull-left[ ```r require("tmap") tm_shape(kde) + tm_polygons(col = "kde_value", palette = "viridis", title = "KDE Estimate") + tm_shape(bars) + tm_bubbles(size = 0.1, col = "red") ``` ] .pull-right[ 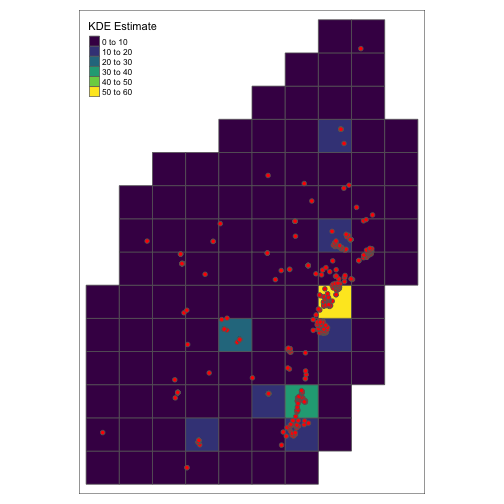<!-- --> ] --- ```r sessionInfo() ``` ``` ## R version 4.2.0 (2022-04-22) ## Platform: x86_64-apple-darwin17.0 (64-bit) ## Running under: macOS Big Sur/Monterey 10.16 ## ## Matrix products: default ## BLAS: /Library/Frameworks/R.framework/Versions/4.2/Resources/lib/libRblas.0.dylib ## LAPACK: /Library/Frameworks/R.framework/Versions/4.2/Resources/lib/libRlapack.dylib ## ## locale: ## [1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8 ## ## attached base packages: ## [1] stats graphics grDevices utils datasets methods base ## ## other attached packages: ## [1] tmap_3.3-3 SpatialKDE_0.7.0 spatstat_2.3-4 ## [4] spatstat.linnet_2.3-2 spatstat.core_2.4-4 rpart_4.1.16 ## [7] nlme_3.1-157 spatstat.random_2.2-0 spatstat.geom_2.4-0 ## [10] spatstat.data_2.2-0 here_1.0.1 sf_1.0-7 ## [13] scales_1.2.0 plotly_4.10.0 lubridate_1.8.0 ## [16] kableExtra_1.3.4 nomnoml_0.2.5 knitr_1.39 ## [19] forcats_0.5.1 stringr_1.4.0 dplyr_1.0.9 ## [22] purrr_0.3.4 readr_2.1.2 tidyr_1.2.0 ## [25] tibble_3.1.7 ggplot2_3.3.6 tidyverse_1.3.1 ## [28] xaringanExtra_0.5.5 xaringanthemer_0.4.1 ## ## loaded via a namespace (and not attached): ## [1] leafem_0.2.0 colorspace_2.0-3 deldir_1.0-6 ## [4] ellipsis_0.3.2 class_7.3-20 leaflet_2.1.1 ## [7] rprojroot_2.0.3 base64enc_0.1-3 fs_1.5.2 ## [10] dichromat_2.0-0.1 rstudioapi_0.13 proxy_0.4-26 ## [13] farver_2.1.0 xaringan_0.24 fansi_1.0.3 ## [16] xml2_1.3.3 splines_4.2.0 codetools_0.2-18 ## [19] polyclip_1.10-0 jsonlite_1.8.0 tmaptools_3.1-1 ## [22] broom_0.8.0 dbplyr_2.1.1 png_0.1-7 ## [25] spatstat.sparse_2.1-1 compiler_4.2.0 httr_1.4.3 ## [28] backports_1.4.1 assertthat_0.2.1 Matrix_1.4-1 ## [31] fastmap_1.1.0 lazyeval_0.2.2 cli_3.3.0 ## [34] s2_1.0.7 htmltools_0.5.2 tools_4.2.0 ## [37] gtable_0.3.0 glue_1.6.2 wk_0.6.0 ## [40] Rcpp_1.0.8.3 raster_3.5-15 cellranger_1.1.0 ## [43] jquerylib_0.1.4 vctrs_0.4.1 svglite_2.1.0 ## [46] leafsync_0.1.0 crosstalk_1.2.0 lwgeom_0.2-8 ## [49] xfun_0.30 rvest_1.0.2 lifecycle_1.0.1 ## [52] XML_3.99-0.10 goftest_1.2-3 terra_1.5-21 ## [55] MASS_7.3-56 hms_1.1.1 spatstat.utils_2.3-1 ## [58] parallel_4.2.0 RColorBrewer_1.1-3 yaml_2.3.5 ## [61] sass_0.4.1 stringi_1.7.6 highr_0.9 ## [64] e1071_1.7-9 rlang_1.0.2 pkgconfig_2.0.3 ## [67] systemfonts_1.0.4 evaluate_0.15 lattice_0.20-45 ## [70] tensor_1.5 labeling_0.4.2 htmlwidgets_1.5.4 ## [73] tidyselect_1.1.2 magrittr_2.0.3 R6_2.5.1 ## [76] generics_0.1.2 DBI_1.1.2 pillar_1.7.0 ## [79] haven_2.5.0 whisker_0.4 withr_2.5.0 ## [82] mgcv_1.8-40 stars_0.5-5 units_0.8-0 ## [85] sp_1.4-7 abind_1.4-5 modelr_0.1.8 ## [88] crayon_1.5.1 KernSmooth_2.23-20 utf8_1.2.2 ## [91] tzdb_0.3.0 rmarkdown_2.14 isoband_0.2.5 ## [94] grid_4.2.0 readxl_1.4.0 data.table_1.14.2 ## [97] reprex_2.0.1 digest_0.6.29 classInt_0.4-3 ## [100] webshot_0.5.3 munsell_0.5.0 viridisLite_0.4.0 ## [103] bslib_0.3.1 ```